At present MOLGEN-MS is available for MS Windows 95/98/NT4.0. In order to use MOLGEN-MS the following hardware requirements have to be fulfilled:
You need an IBM-compatible PC (80486 or higher) running MS Windows 95, Windows 98 or Windows NT 4.0.
A 3.5" disc drive or a CD-ROM drive must be available for installation.
A mouse is required as some parts of the program can be used exclusively by a mouse.
On the hard disk you need about 10.0 MB storage space. The space which is needed for a run depends of course on the particular elucidation problem, i.e. on the number of generated candidates for the structural formula. This size depends on the molecular mass, the candidates for the molecular formula and the prescribed and forbidden substructures
If all these requirements are fulfilled you may now start installing MOLGEN-MS. To do this, carry out the following steps:
Insert the MOLGEN-MS installation disk/CD-ROM into your floppy disk/CD-ROM drive.
Open the EXPLORER and click on the 3,5 floppy/CD-ROM folder. Then double-click on the Setup program icon to start the installation. Follow the instructions of the MOLGEN-MS installation program.
During the installation the user is asked for an install directory. By default this directory has the name MOLGEN-MS. For a proper installation be aware that there are no blanks in the pathname of this folder.
The setup program copies several subdirectories into this folder as there are:
When the installation is finished, MOLGEN-MS is ready to start and you can execute it by double-clicking on the MOLGEN-MS icon on the desktop.
At the first start of MOLGEN-MS the following window will appear:

Now enter your license number, which you should have got with your personal copy of MOLGEN-MS and click Enter. Then your copy of MOLGEN-MS will be licensed for this computer. The MOLGEN-MS-program cannot be copied to another computer since it will work only on the machine it is licensed for.
If you do not have a valid license number, MOLGEN-MS will only run as limited demo version. Then at most 6 structures will be shown in the structure viewer and you can only open the spectra file named tra-demo.tra, which is distributed with every copy of MOLGEN-MS.
You are now ready for your first structure elucidation supported by MOLGEN-MS.
In order to open the mass spectra file tra-demo.tra you have to proceed as follows:
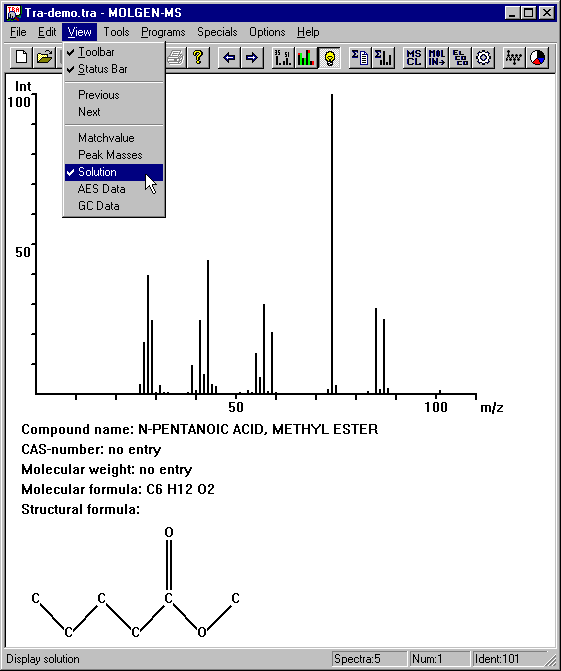
Now you can see the first mass spectrum in the MOLGEN-MS main window:
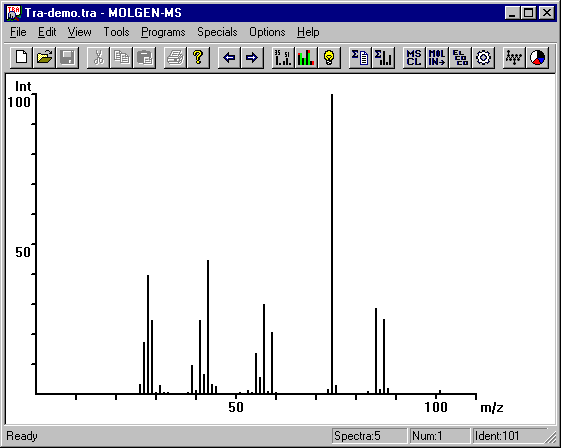
In the title you see the name of the spectra file: Tra-demo.tra. In the lower right corner you find additional data about the file: It contains 5 spectra, the displayed one is the first in the file and its Identification No. is 101.
Select the menu item Programs|Expert Mode to get to the Expert Mode-dialogue.
You see the settings for the single steps of the interpretation. The meaning of these parameters will be explained later on. Click the Start-button to begin the automatic structure elucidation. After a little while the window will look as follows:
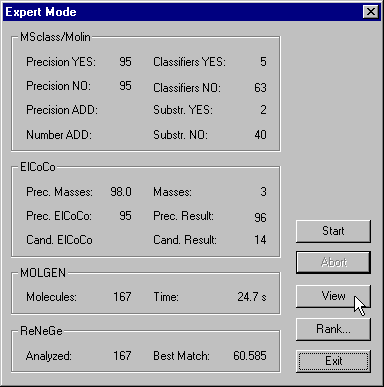
There were 167 molecular graphs generated as proposals for the structural formula of the compound in question and the best one matches 60.585% of the spectra's total ion current (TIC)
Of course, you may want to see the result of the computation now. For this purpose, select the View-button and obtain the desired visualization of the generated structural formulae:
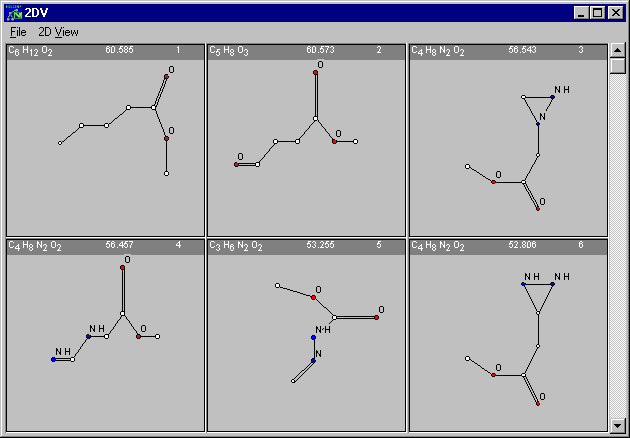
Are you able to find out the one that belongs to the spectrum in question ? If you are not sure, close the Expert Mode-dialogue by clicking Exit and choose View|Solution. Some additional data that is stored in the tra-demo.tra file will appear in the MOLGEN-MS main window, including the structural formula of the compound in question.
Apart from the expert mode, the user can access each step of the whole system on its own. The settings of each program can be changed. If misinterpretations happen during the program run, their reasons can be explained. Questions like "What happened if ..." can be discussed and systematic structure elucidation can be executed at a high level of usability.
Classification of the displayed mass spectrum is executed as follows: Click Programs|MSclass and the MSclass dialogue appears.
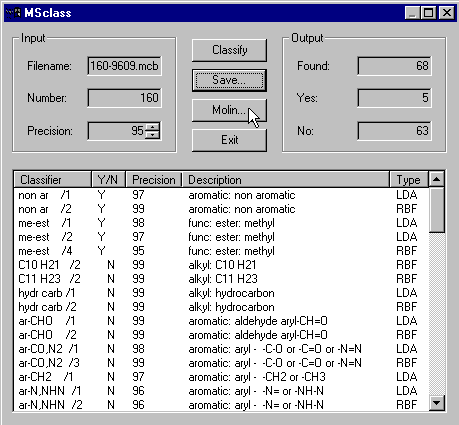
By default the program applies the classifier file 160-9609.mcb which contains 160 classifiers. Default precision is 90%. For this example there were 68 classifier answers with precision at least 95% found (5 times YES and 63 times No).
In the dialogue's listbox you find detailed information about the found classifiers: the classifier names, their boolean answer, the precision for this answer, the value of the discriminant variable, a description of the structural property (SP) treated by the classifier and the type of classification method. By clicking the column headers you can change sort the classifiers by their attributes. For instance if you want them to be sorted by their description, press the Description-button. If you click the classifier names with the right mouse button, a graphical description of the classifier is displayed. For me-est /1 you get
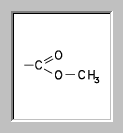
Perhaps you already recognized that three classifiers with description func: ester: methyl answered with YES. Therefore we can assume that the unknown has a functional group methyl ester as substructure. How this information will be processed is explained in the next step.
Click the Molin button to get to the next dialog, which is dedicated to the selection and conversion of the classification results into topological information for the structure generator.
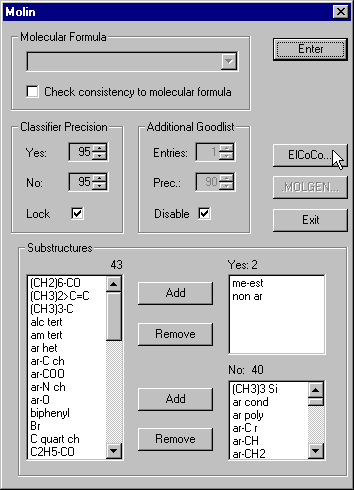
Again, there is the possibility to enter the lower border for the classifier precision. By default this border is 95% for both, YES and No answers. There are three list boxes for the SPs that can be recognized by MSclass: One for the SPs that must occur in the molecule, one for forbidden SPs and one for the remaining SPs for which no classifier answers were found. In the YES listbox we have me-est which stands for a functional group methyl ester. A SP is added to the Yes or No listbox if at least one classifier for this substructure answers with the required precision. The SPs for which no classifiers were found are displayed in the left listbox. Even if we have one YES and one NO the SP will be found in the left listbox.
If the molecular formula is known it can be entered and substructures that are not consistent can be removed from the YES and NO listboxes automatically. If the molecular formula is not known, there is an algorithm that finds most likely candidates for the elemental composition.
Click the the ElCoCo button to proceed to the dialog that supports the computations concerning the molecular formula.
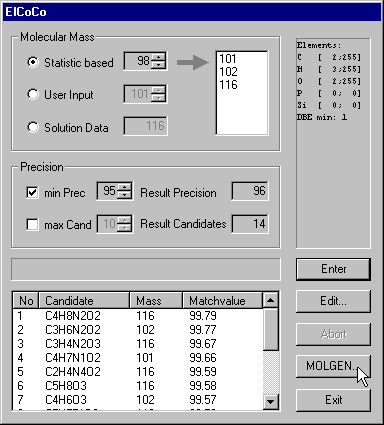
Let's have a closer look at the controls. The first decision concerns the choice of the molecular mass. As the molecular ion peak is not visible in some electron impact mass spectra we have to consider several candidates for the molecular mass. In our example these are 101, 102 and 116. 101 is the mass of the highest peak of the first peakgroup. The other proposals 102, 112 result from statistical considerations.
The prescribed substructure me-est induces certain intervals for the numbers of atoms of each chemical element. As me-est consists of two carbon, three hydrogen and two oxygen atoms, the molecular formula must have at least 2 C, 3 H and 2 O atoms. The upper bounds 255 are default settings. The intervals 0-0 for P and Si result from classifiers concerning the presence or absence of elements P and Si. As in the me-est substructure there is a double bond, the molecular formula must have at least one double bond equivalent.
Even ElCoCo needs a minimum precision as input. 95% can be understood in the following way: you want to have the correct molecular formula with probability 95% among the computed candidates. A higher precision will cause more candidates. Of course these percentages are only rough estimations and the actual behavior depends very much on the quality of your experimental data.
Now click the enter button to start the computation. This can take several minutes and the progress bar tells you how the computation proceeds. When it's finished, the candidates are displayed, ordered by increasing matchvalues. The bigger the matchvalue is, the better the experimental spectrum can be explained. More precisely the matchvalue tells, how much percent of the TIC can be explained by the algorithm. Therefore, the matchvalue is always a number between 0 and 100.
In this example we have 14 candidates for the molecular formula. The next step is to compute all connectivity-isomers to the molecular formulae proposed by ElCoCo which fullfil the substructure restrictions obtained form MSclass.
By clicking the MOLGEN-button the MOLGEN-dialogue appears.
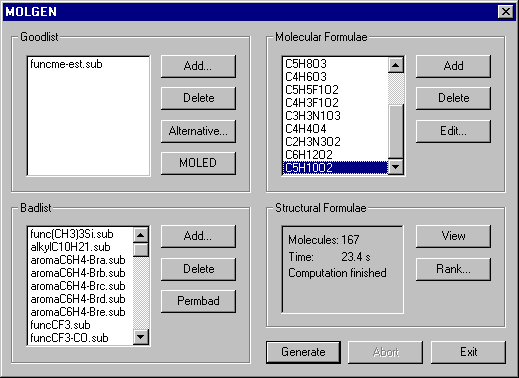
In the Goodlist and the Badlist we have the filenames of the MOLGEN substructure files. In the right listbox there are the molecular formulae suggested by ElCoCo. You start the structure generation by clicking the Generate-button. Now for each of the listed molecular formulae all connectivity-isomers are generated that fullfil the structural restrictions. The generation is redundancy free and exhaustive. In this example we have a total of 167 structural formulae. If you simply like view them, click the View-button. But MOLGEN-MS offers a further algorithm, that allows to rank the candidate structures with decreasing likelihood to be the structure formula of the unknown.
Therefore click the Rank-button, that leads you to the ReNeGe-dialogue. When the window appears, the generation of the MS reaction networks is already started. On the left there are two listboxes with the available reaction mechanisms.Only checked reactions are used for the generation of the reaction networks. In the central listbox you find detailed information about the structure candidates: Their logical number, their molecular formulae and their molecular weight.
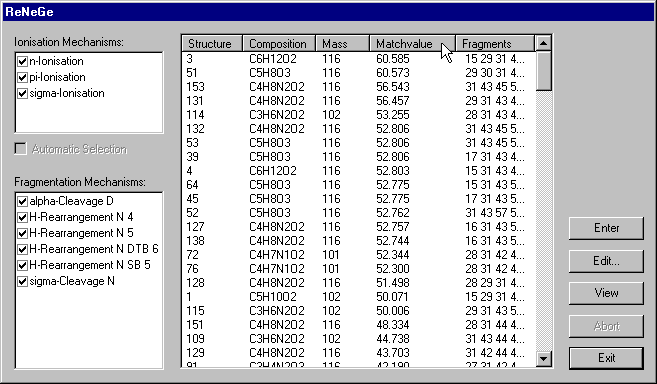
The last two columns carry information about the results of the ranking. In the last column, there is a list of all weights of possible fragments, and in the column with the header Matchvalue there is a number that tells how much percent of the experimental spectra's TIC can be explained by virtual fragments.
When you click a column header, the structures are sorted by the corresponding attribute. So if you press the Matchvalue-button, the structures become ordered by decreasing matchvalues.Selecting a structure with the right mouse button opens a window with the structure's properties including a drawing :
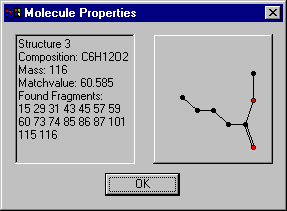
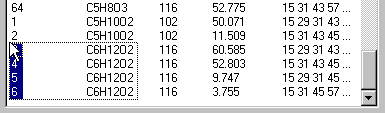
The Structure-listbox is a multi-select listcontrol, i.e. you can select several items with the left mouse button and the shift- and strg-key. For instance if you want to see all structures with molecular formula C6H12O2, click the Composition-button to sort the molecules by their elemental composition and select those with the desired molecular formula:
Now press the View-button and the selected structures will be displayed
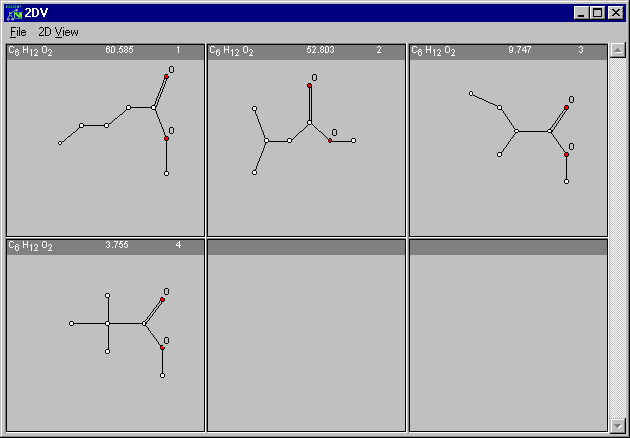
If you choose View without having any structures selected, all molecules will be shown.